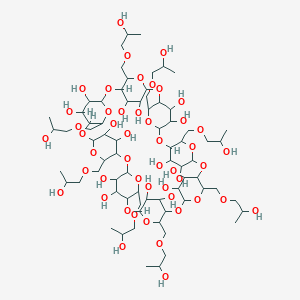
Details of the Drug Inactive Ingredient (DIG)
| Full List of Biological Targets of DIG (DBTs) Regulated by This DIG | ||||||
|---|---|---|---|---|---|---|
| Oxidoreductase (ORase) | ||||||
| DBT Name: Monoamine oxidase A (MAO-A) | Click to Show/Hide | |||||
| Detailed Information |
DBT Info
 click to show the detail info of this DBT click to show the detail info of this DBT
|
|||||
| Experiment for Assessing the Biological Activity of the Studied DIG on This DBT | ||||||
| Biological Activity | IC50 = 167 nM (estimated based on the structural similarity with CHEMBL1629810 ) | [1] | ||||
| Structural Similarity | Tanimoto coefficient = 1 | |||||
| Tested Species | Homo sapiens (Human) | |||||
| UniProt ID | AOFA_HUMAN | |||||
| Potential-driven transporter (PDT) | ||||||
| DBT Name: Dopamine transporter (DAT) | Click to Show/Hide | |||||
| Detailed Information |
DBT Info
 click to show the detail info of this DBT click to show the detail info of this DBT
|
|||||
| Experiment for Assessing the Biological Activity of the Studied DIG on This DBT | ||||||
| Biological Activity | IC50 = 1687 nM (estimated based on the structural similarity with CHEMBL1629810 ) | [1] | ||||
| Structural Similarity | Tanimoto coefficient = 1 | |||||
| Tested Species | Homo sapiens (Human) | |||||
| UniProt ID | SC6A3_HUMAN | |||||
| References | |||||
|---|---|---|---|---|---|
| 1 | DrugMatrix in vitro pharmacology data. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Zhang and Dr. Mou.